SINGEK BLOG: Tadpoles experiencing mass die-offs: Are protozoan parasites the cause?
 As recently as 2006, travellers to Panama might have been treated to the spectacle of golden frogs brilliantly coloured against the deep green leafy background of the high mountains. Or perhaps they would have heard their calls rising up in a chorus with numbers reaching the thousands.
As recently as 2006, travellers to Panama might have been treated to the spectacle of golden frogs brilliantly coloured against the deep green leafy background of the high mountains. Or perhaps they would have heard their calls rising up in a chorus with numbers reaching the thousands.
Today these frogs have disappeared from the wild and can only be found in small captive breeding colonies. The principle cause of their decline has been identified as the pathogenic chytrid fungus Batrachochytrium dendrobatidis (unwittingly spread by the same travellers that used to enjoy their spectacle).
Unfortunately, the plight of amphibians extends well beyond Panama. Nearly one-third of known species worldwide are threatened, while an additional 42 percent are reported as ‘declining in population’ (http://www.iucnredlist.org/initiatives/amphibians/analysis; accessed June 15, 2017). Like the Panamanian golden frog, many are imperilled by infectious disease.
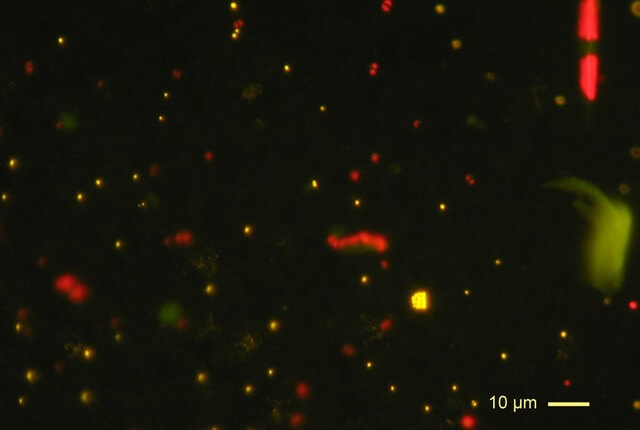
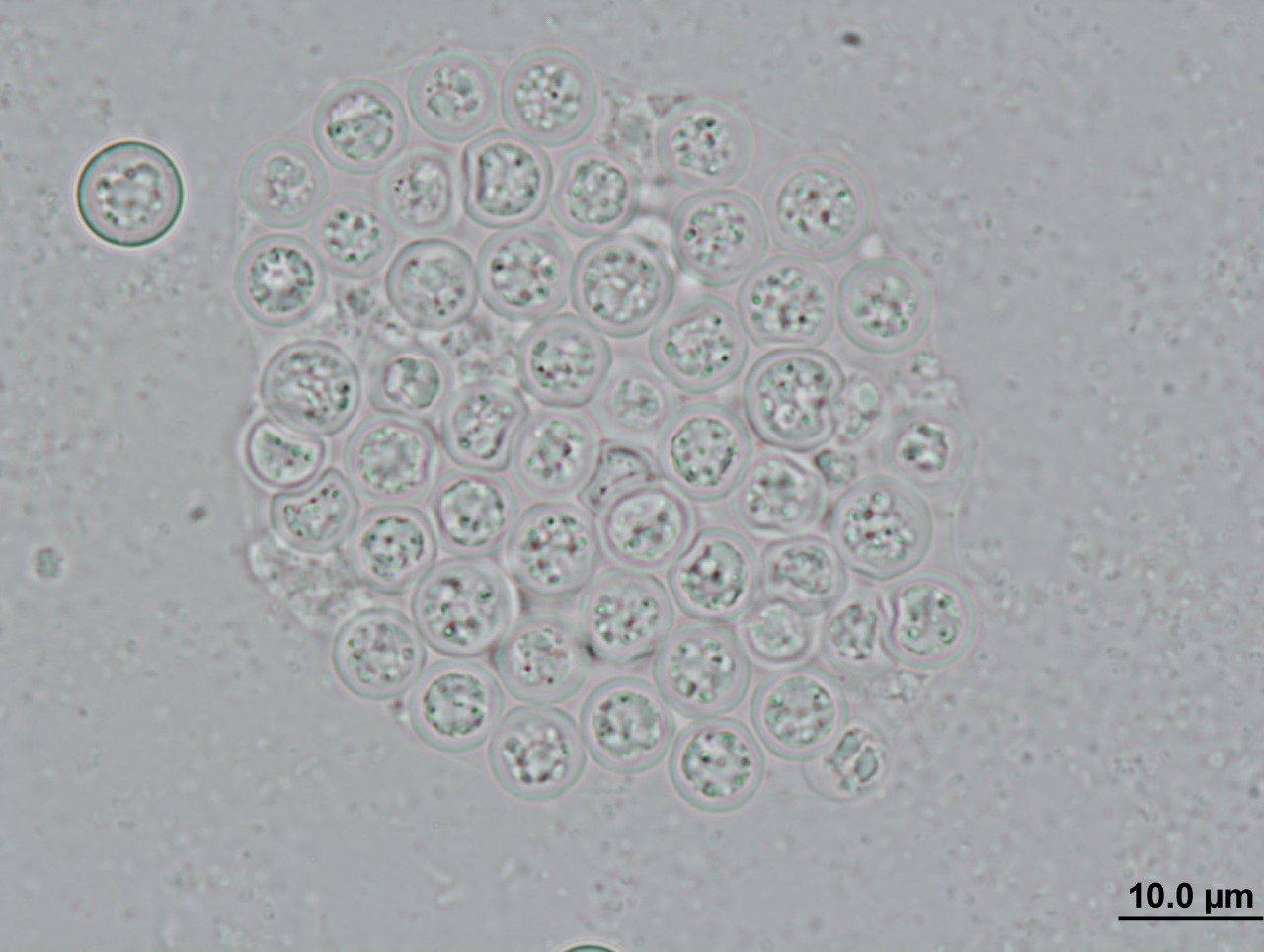
About the same time as the golden frog’s disappearance, researchers observed a mass die-off of southern leopard frog tadpoles from several ponds in Georgia, USA (Davis et al. 2007). They discovered that their body tissues, particularly their livers, had been infiltrated by a single-celled parasite similar to those in the genus Perkinsus (distant relatives of the agent of malaria) in both their appearance and genetic material.
Interestingly, another group of researchers later discovered Perkinsus-like genetic material in the tissues of tadpoles from 14 genera across a broad geographic range (Chambouvet et al. 2015); however, none of these tadpoles exhibited the symptoms that devastated the populations in Georgia. Indeed, it seems that the parasite can also cryptically infect amphibians without causing disease.
The outcome of infection is most likely determined by a complex interplay between the genes of the host and parasite, parasite levels in the environment, and extrinsic environmental factors. Over the next few years, my colleagues and I will begin to tease apart these dynamics.
To first establish disease causation, we will conduct infection experiments using different amphibian host species, inoculation methods and parasite burdens. These experiments will be followed by RNA analysis to identify the genes used by the parasite to facilitate infection. We also plan to conduct field studies of localities with reported die-offs to survey parasite levels and conditions that might be conducive to their pathogenesis.
Finally, we will use drug-sensitivity screening to identify compounds that inhibit growth of the parasite, providing a means to safeguard a captive breeding population should another species share the fate of the Panamanian golden frog.
About the author
Vanessa Smilansky – ESR 11
 My MSc research was conducted at the University at Buffalo in Buffalo, NY, USA. It involved the characterization of population genetic structure in a surface brooding Caribbean octocoral using microsatellite genotyping, genetic distance measurement and spatial autocorrelation. The research aimed to elucidate larval dispersal patterns, which we concluded occur on a fine-scale that varies temporally.
My MSc research was conducted at the University at Buffalo in Buffalo, NY, USA. It involved the characterization of population genetic structure in a surface brooding Caribbean octocoral using microsatellite genotyping, genetic distance measurement and spatial autocorrelation. The research aimed to elucidate larval dispersal patterns, which we concluded occur on a fine-scale that varies temporally.
Following the completion of my degree in 2013, I worked as a genetics technician in the Washington Department of Fish and Wildlife in Olympia, WA, USA. My work involved genotyping salmonids with microsatellite and SNP markers for use in genetic stock identification and parentage analysis. Similar to the structure of the SINGEK network, the complexity of our projects and their far-reaching impacts led to a collaborative effort with other professional organisations both within and outside the agency.