SINGEK BLOG: The Virus in a drop of ocean and in a drop of you
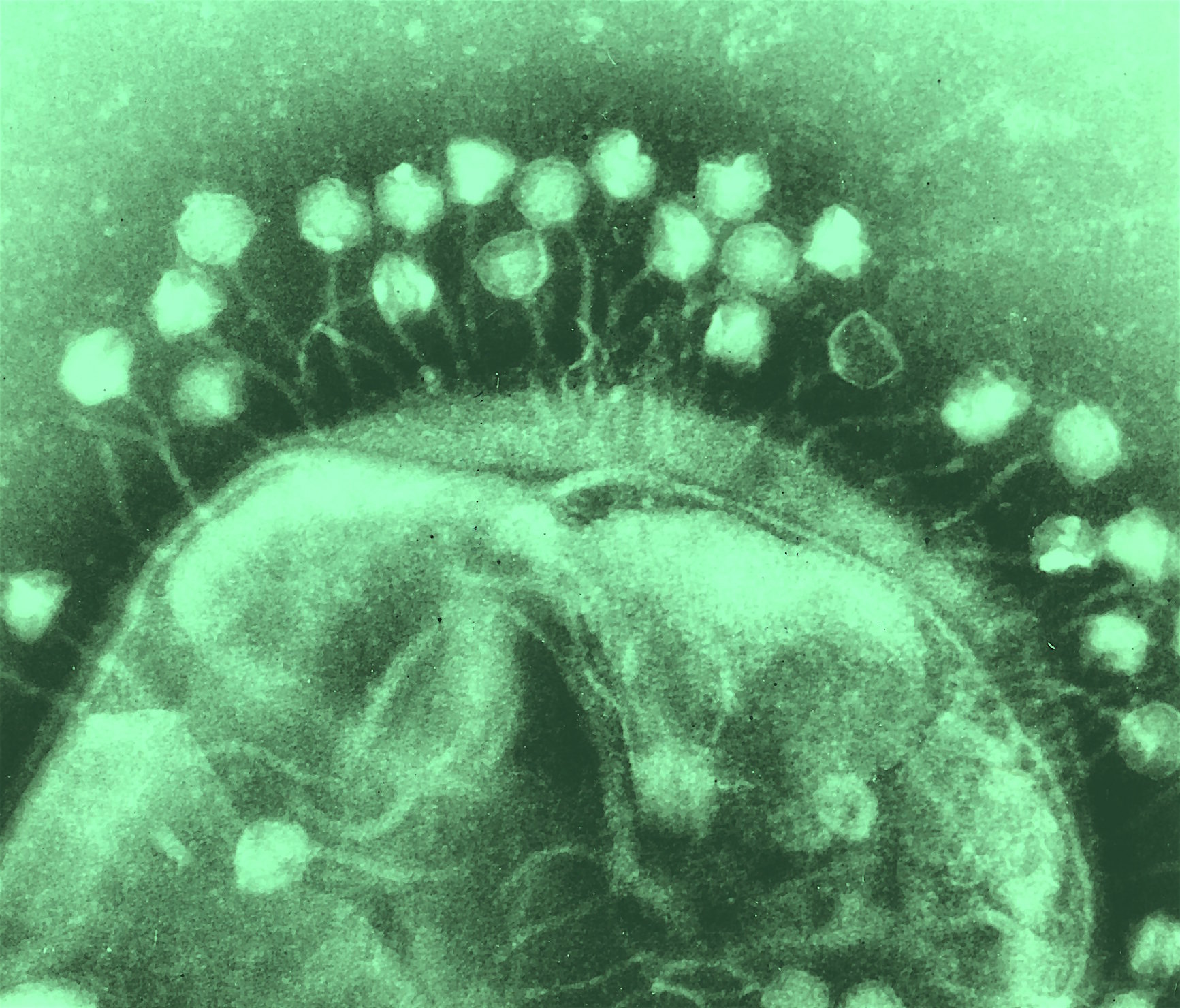
Electron micrograph of bacteriophages (bacterial viruses) attached to a host cell. Image modified from original by Dr Graham Beards CC BY-SA 3.0
Viruses. They are everywhere. They are the most abundant creatures on the planet. Probably you have heard about them when you get sick at home for a few days or when you watch the news about the next big pandemic. Now, in your body could be around thousands of viruses in some cells of you. Most certainly you don’t like them at all, but give viruses a chance. If there were no viruses, life on our planet would be very different, and maybe you and everyone you know and care about would not exist.
We will back into you later, because now I want you to consider a drop of ocean. In every liter of marine water there is about a billion of viral particles. In fact, in each wave there might be more viruses than there are visible stars in the sky.
But viruses can’t exist by themselves. In order to be active and replicate, that is, produce more viruses, they have to infect a living cell. So in a droplet of marine water there is a billion year old war between cellular organisms and viral organisms. This “microbial war” has profound impacts, for example on our planet’s climate. Some viruses infect algal cells, that is tiny plant-like organisms which also use sunshine to grow. They cause the algae to modify the production of some gases which help to create clouds above the ocean and other gases, such as CO2 which has a significant role in climate change.
There are marine viruses which can even stimulate food production from sunlight, technically called “photosynthesis”, by infecting some kinds of microbes. Some small viruses, called “virophages” help their hosts protecting against other giant viruses. And ultimately, some can even go deeper than that by becoming one with their hosts. By integrating their genetic material into the host DNA, they are blurring the boundary between virus and host cell.
I hope I had convinced you by now that there is much more about viruses than diseases. But how do viruses connect to your own existence? Well, there is a remarkable similarity between proteins that make up the placenta (the structure that protects the embryo inside the womb) and viral envelope proteins. This similarity is unlikely to be due to chance alone. Scientists hypothesise that around 150 million years ago, an integration event of viral DNA into the genome of a mammalian ancestor of ours has occurred, giving rise to the placenta in its current form.
This entangled history between viruses and cells could have allowed placentarians to become the dominant form of life, creating arts, politics, science and controlling our planet. But now that you know more about the viral world and their roles, you may start to ask yourself who really is in control, and if there is a single winner in this billion-year old planetary war, that is still happening right now, in a drop of sea water, or in every drop of you.
About the author

Luiz Felipe de Almeida – ESR 9
I am a Biologist from Brazil (UNISINOS), Master in “Biodiversity and Evolutionary Biology” (UFRJ) and currently a PhD candidate from Université Pierre et Marie Curie – Sorbonne in the Observatoire Océanologique Banyuls-sur-mer working in the group “GENOPHY” with marine microalgae and its bacterial and viral interactions. My SINGEK ESR 9 research project is “Genomic insights into green microalgae and their interactions with viruses and bacteria”.

 JAVIER FLORENZA – ESR 4
JAVIER FLORENZA – ESR 4