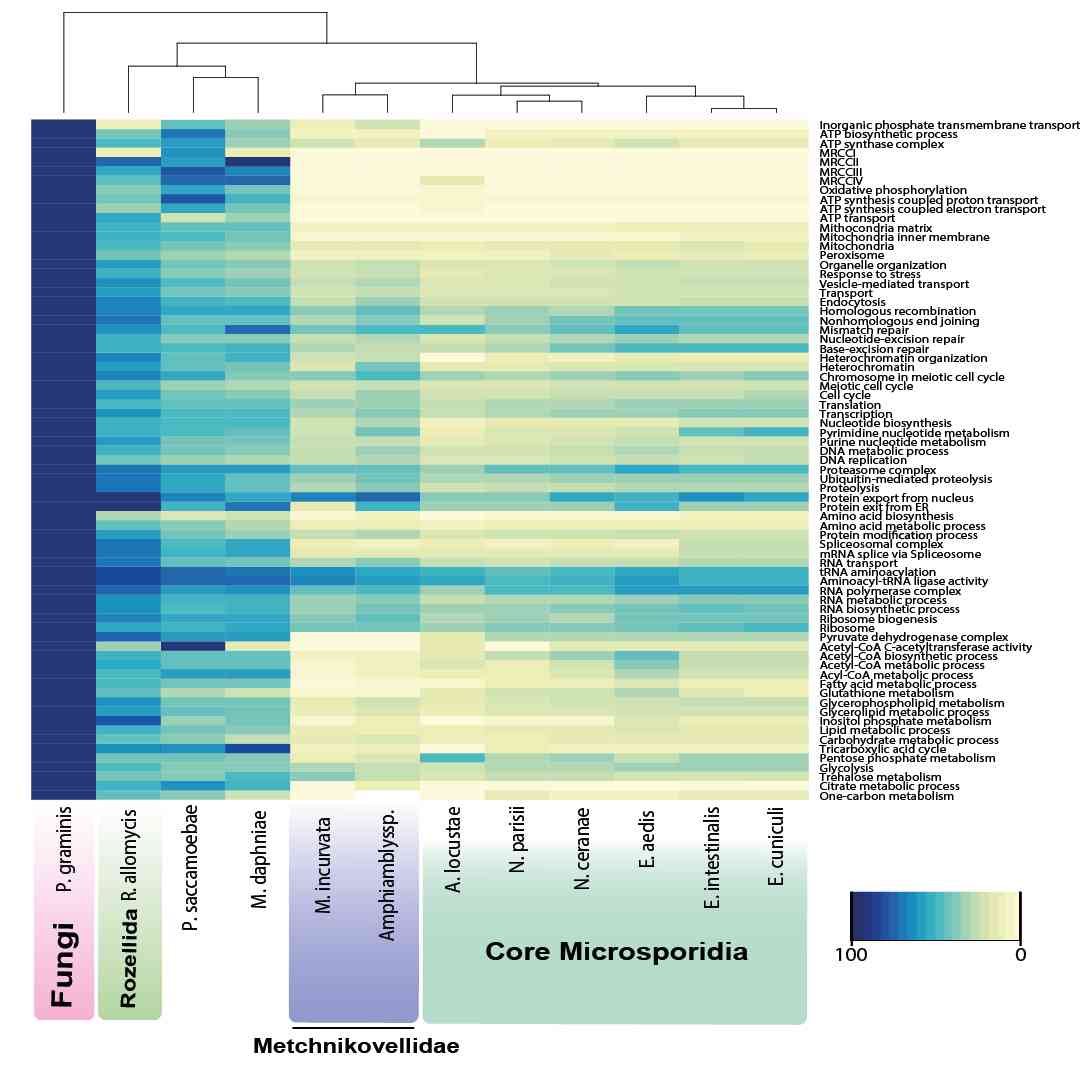
A collaboration between different SINGEK teams from the Ghent University (VIB) and the Observatoire Oceanologique de Banyuls sur mer (OBS Banyuls-CNRS) has been established. Within this context, François BUCCHINI (SINGEK ESR10) has developed code to assess the gene space completeness in the newly sequenced Picochlorum genome and compared it with genomes of other green algae. From that, it could be concluded that the new genome was of high quality and more complete than published Picochlorum genomes.
As a result of this analysis, a paper has recently been published in Genome Biology and Evolution:
M. Krasovec, E. Vancaester, S. Rombauts, F. Bucchini, S. Yau, C. Hemon, H. Lebredonchel, N. Grimsley, H. Moreau, S. Sanchez-Brosseau, K. Vandepoele, G. Piganeau
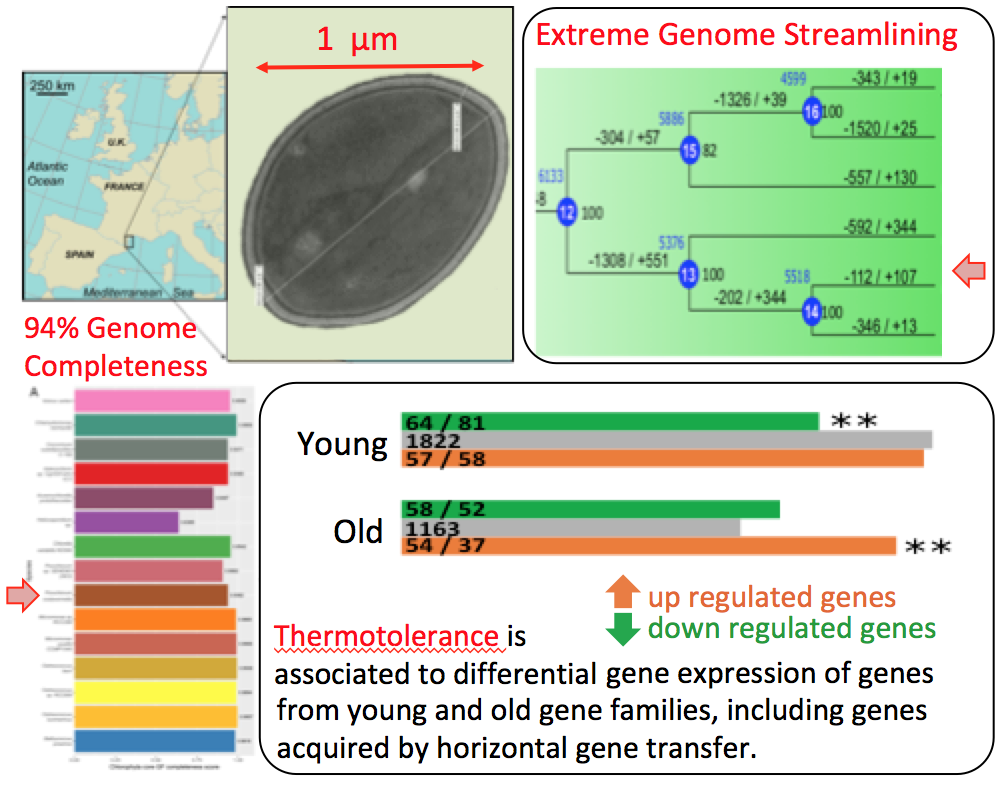
The Genome of a novel picoalga species, Picochlorum costavermella, provides insights into the Evolution of Thermotolerance in the Green lineage.
While the molecular events involved in cell responses to heat stress have been extensively studied, our understanding of the genetic basis of basal thermotolerance, and particularly its evolution within the green lineage, remains limited. Here, we present the 13.3-Mb haploid genome and transcriptomes of a halotolerant and thermotolerant unicellular green alga, Picochlorum costavermella (Trebouxiophyceae) to investigate the evolution of the genomic basis of thermotolerance. Differential gene expression at high and standard temperatures revealed that more of the gene families containing up-regulated genes at high temperature were recently evolved, and less originated at the ancestor of green plants. Inversely, there was an excess of ancient gene families containing transcriptionally repressed genes. Interestingly, there is a striking overlap between the thermotolerance and halotolerance transcriptional rewiring, as more than one-third of the gene families up-regulated at 35 °C were also up-regulated under variable salt concentrations in Picochlorum SE3. Moreover, phylogenetic analysis of the 9,304 protein coding genes revealed 26 genes of horizontally transferred origin in P. costavermella, of which five were differentially expressed at higher temperature. Altogether, these results provide new insights about how the genomic basis of adaptation to halo- and thermotolerance evolved in the green lineage.