Background
Microbial eukaryotes increasingly attract the interest of various scientific disciplines. These microbes encompass the majority of eukaryotic diversity and are critical for understanding the evolution and diversification of the eukaryotic form, including the origin of plants and animals. As primary producers, grazers and decomposers, microbial eukaryotes play key roles in ecosystem functioning. They frequently establish close interactions with other organisms, from parasitism to mutualism, with critical ecosystemic, socio-economic or biomedical impacts.
Despite their importance, the vast majority of microbial diversity is currently uncultured and therefore intractable for investigation, specifically preventing traditional genomics analysis based on obtaining high biomass of pure microbial cultures prior to sequencing. This knowledge gap has been referred to as the “dark matter” of biodiversity.
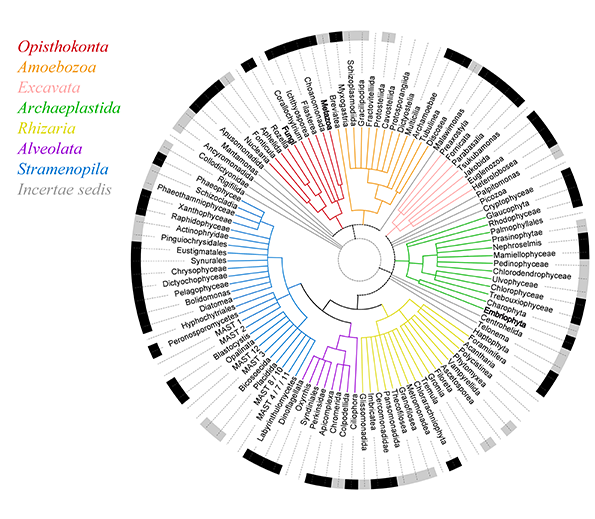
Figure 1. Our current vision of the Eukaryotic Tree of Life. Taxonomic groups, coloured according to their adscription to supergroups, are noted in the external circle as having genomic/transcriptomic data (in black), having cultures but no genomic data (in grey), or being uncultured (in white). Arguably only groups in black can be assumed to be well characterized, highlighting our incomplete view of eukaryotic diversity. Source: del Campo et al. 2014.
Single cell genomics (SCG) is an emerging technology in biological sciences that has recently become feasible due to the improvements in single cell manipulation, whole genome amplification, high-throughput sequencing and bioinformatics. Now, for the first time, these tools enable the sequencing of genomes of individual cells. Single cell genomic exploration of the plethora of ecologically dominant but elusive microbial eukaryotes offer tremendous promises to advance in multiple fundamental and applied research fields, including ecology, evolution, and biotechnology, and is about to become one of the most dominant molecular techniques given appropriate development.
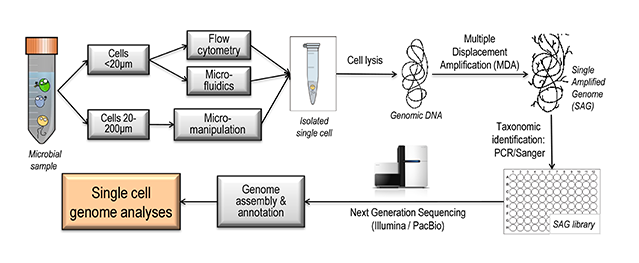
Figure 2. Overview of the steps involved in Single Cell Genomics
Applying SCG to microbial eukaryotes is still in its infancy, and is particularly challenging when compared to SCG on prokaryotes, because of target genomes of greater complexity (multiple chromosomes, introns, low codon density, high numbers of repeats, large gene paralogue families), and the need to combine SCG with transcriptomics (sequencing messenger RNA). Despite these difficulties, analysing genomes/ transcriptomes of single microeukaryotic cells is crucial for two main reasons:
- De novo genomes from uncultured lineages provide the means to address pertinent ecological and evolutionary questions such as: What is the true biodiversity of microbial eukaryotic assemblages? How does the uncultured diversity populate the eukaryotic tree of life, including novel branches and deep evolutionary events? Can genetic repertoires explain the success of certain uncultured lineages in natural environments? Can genomic data explain the functional diversity of related lineages occupying similar ecological fields? What is the distribution in the ocean of species and functions, and what are their drivers?
- SCG allows a view of the genome variability within a microbial population. This is used in biomedicine to understand cancer origins and development and can also be applied in classical population genetics that aims to describe genetic variation accumulation within populations prior to diversification, or to understand how microbial cells interact and cooperate to perform key ecological functions. SCG can shed light on diversity and evolutionary questions such as: Which interactions are established between single eukaryotic cells of given lineages and other eukaryote species, prokaryotes and viruses, and what are their ecological implications (infection, cooperation, specific grazing)? What is the genetic structure of microbial eukaryote populations and how do they distribute in the spatio-temporal space? How do genomic changes underpin diversification in microbial eukaryotes?
Objectives
The main objective of the SINGEK ITN project is to explore SCG of microbial eukaryotes to its full potential by training a new generation of researchers and do so by addressing a set of fundamental and inter-related scientific questions. All Early Stage Researchers (ESRs) will experience all steps in the process encompassing complementary technological and scientific backgrounds (laboratory, bioinformatics, ecology and evolution). Each ESR will contribute to the Consortium by devoting most of his/her time to a specific research goal, and provide state-of-the-art technological and scientific advances that will feed the central theme of the Consortium: advance in the SCG of microbial eukaryotes for evolutionary and ecological research. The ESR integration in the planned research will be accomplished by collegiate scientific direction, training events and courses of transferable skills, and secondments to basic and applied research institutes and private enterprises.
Work Packages
The activities planned in SINGEK are structured in a set of work packages that include management (WP1: Project Management and coordination), training (WP2: Network-wide training activities), technological developments (WP3: Laboratory work, from single cells to sequences and WP4: Bioinformatics), scientific assessments (WP5: Application of SCG in microbial ecology and WP6: Application of SCG in microbial evolution) and dissemination (WP7: Dissemination and communication). The core of the research programme (WP3 to WP6) covers all aspects of the SCG pipeline.
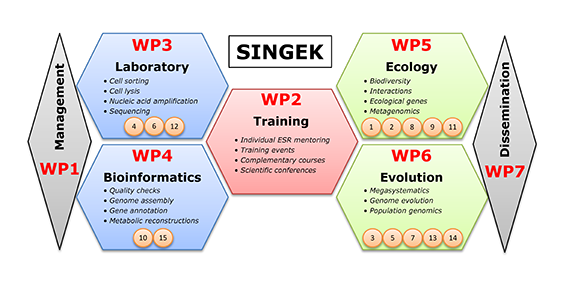
Figure 3. Schematic representation of the main activities planned for the SINGEK project. The specific research of each ESR (noted as orange circles) relates to the methodological (WP3/WP4) or scientific work packages (WP5/WP6).
The first research work package comprises laboratory work (WP3), which includes single cell sorting, nucleic acid amplification from the minute quantities in a single cell, and highthroughput sequencing. The second is bioinformatics (WP4), by which the billions of short sequences are processed into comprehensive datasets, requiring assembly into larger contigs, gene prediction and annotation, and reconstruction of metabolic pathways. The generated genomic data is then exploited to address scientific questions within two tightly interconnected disciplines, which if tackled separately could not be answered. Fundamental ecological questions (WP5) include the study of the biodiversity of natural samples, cell-to-cell interactions, and gene complements of uncultured lineages that could explain their lifestyle and ecological function. Evolutionary questions (WP6) include the addition of uncultured lineages to retrieve a more complete eukaryotic Tree of Life, understanding the evolution of genes and pathways across different lineages, and population genomics in a given lineage or species group.