SINGEK BLOG: The Broad Applications of Single Cell Technology
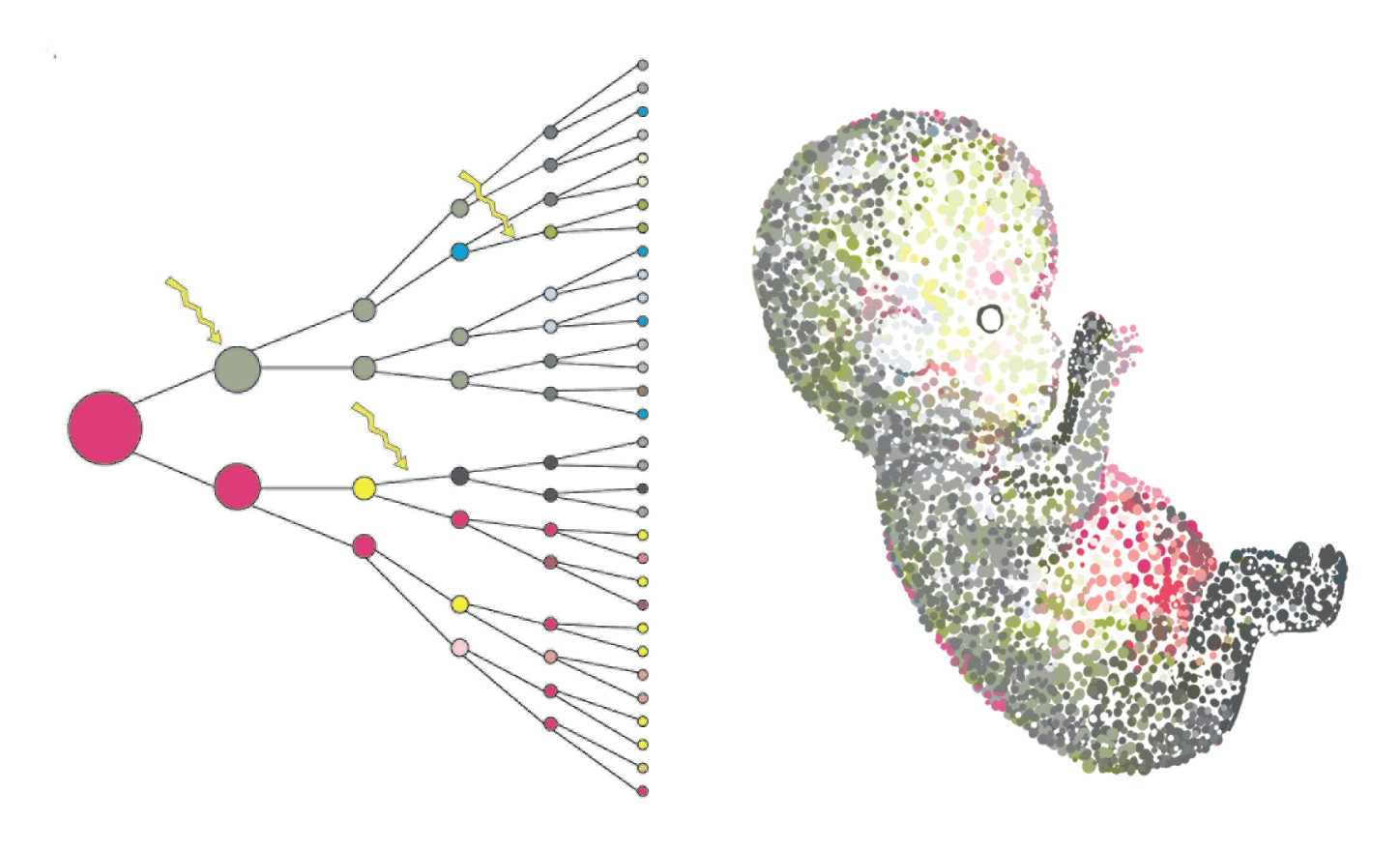
All multicellular life start from a single cell. As cells divide and differentiate , changes in transcriptional profile occur, leading to considerable phenotypical diversities forming different subpopulation of cells and tissues in living organisms.
Figure Credits: Tarryn Porter and Iain Macaulay, Sanger Institute.
Single cell technology – the technology that makes it possible to study individual cells rather studying cultured colonies consisting of thousands or millions of cells – has provided us a magnifier to look at cells one by one and to study them separately and of course more precisely!
So far under the SINGEK consortium, we have always been talking about single cells that are uncultured microbes or more specifically microbial eukaryotes. We have been talking about how these organisms are crucial for understanding evolution and the functioning of ecosystems. We also discussed the best methods to study these single cells, from procedures to collect the samples to the tools to define their position in a phylogenetic tree of life.
Single cell technology is being used beyond SINGEK in many different fields of biological research. Beside Microbiology and Evolution, research areas like Complex tissues, Neurobiology, Immunology and Cancer are among the top fields that single cell technology is being used. In the same ways that we are able to separate microeukaryotic single cell organisms, we are also able to separate and study tissue specific cells, neurons, sperm cells, oocytes and cancer cells. Nowadays many researchers are interested to take advantage of single cell technology to answer questions like:
- How do tissues develop from stem cells? This can be done by tracking the path of individual cells while evolving to specific tissues like liver or muscle.
- How heterogeneous is each tissue in our body which we maybe superficially consider as collection of identical cells?
- How do embryos develop to give rise to a complete multicellular organism with different tissues, each specialized in a specific set of functions?
- How do immune system specific cells react and behave in response to infections and other perturbations?
- How do tumors evolve from individual, regular cells?
All these and many more questions are new approaches that single cell technology provides a reasonable amount of precision to study. The challenge right now is to improve this technology in terms of how to best separate cells, how to extract the DNA and RNA content from them, how to sequence their genetic information and how to analyze this data to answer new questions that this technology has made us able to think about.
As an Early Stage Researcher in the SINGEK project, I will be working on developing statistical and computational methodologies to address these questions. I will try to integrate machine learning algorithms as well as other statistical methods to better understand the data coming out of single cells, to increase the quality of the data and at the end to extract as much information as possible from this basic units of life.
About the author

Atefeh Lafzi – ESR 6
I am currently a PhD student in Biomedicine and work as SINGEK ESR 6 on development of methodology for single cell organism genome analysis in CRG-CNAG, Barcelona. I am interested in searching for procedures and data analysis methodology for single cell studies of species for which no reference genome sequences exist. Establishment of methodology for homogeneous DNA amplification from single cells, de novo genome assembly, RNA analysis and de novo transcriptome assembly are also among the areas I will work on during my PhD.


Leave a Reply
Want to join the discussion?Feel free to contribute!