SINGEK BLOG: Please, Mind The Gap
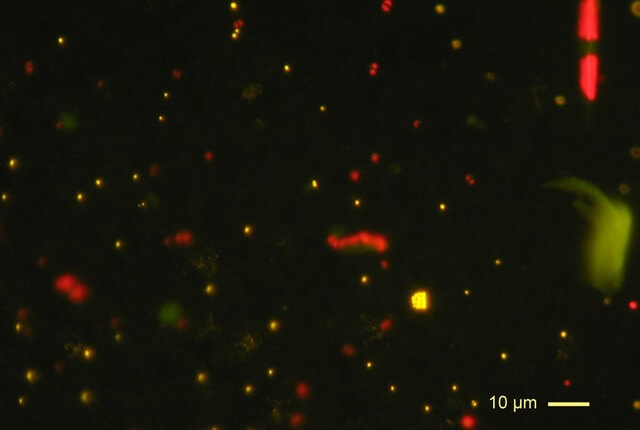
Photosynthetic picoplankton from the Pacific Ocean (off the Marquesas islands observed by epifluorescence microscopy (blue exciting light). Orange fluorescing dots correspond to Synechocococus cyanobacteria, red fluorescing dots to picoeukaryotes. Larger cells (e.g. diatom, upper right) can also be seen. Image original by Daniel Vaulot (CC BY-SA 2.5)
Reporting a lack of genomic diversity in the databases; a call to sequence more reference – and draft-quality genomes to significantly increase the understanding of marine microbial eukaryotes.
The oceans are Earth’s largest ecosystem, representing 71% of the planet’s surface. Despite its vastness, the diversity of the oceans’ life remains rather unknown. In fact, scientists estimate that less than 1% of the total species that it harbors are currently known. Among the lesser-known organisms in the ocean are the protists – single-celled eukaryotic organisms – denoting that they have only one cell, while the term ‘eukaryote’ means that each of these cells has a nucleus (and other organelles). Among the few well studied protists are the Radioloria (beautifully illustrated by Ernst Haeckel) and diatoms, which present a siliceous outer skeleton. But for long time they have often been overlooked and undersampled; they are now considered as a real neglected area of research and the reason are various. Above all: they are hard to study.
Classical taxonomy (the science which studies the classification of life) relies on morphological features of species. And, in spite of the fact that protists can be both plant-like or animal-like and share certain characteristics, they encompass an incredible diversity in their structure and function. However, many of the single cells are morphologically featureless; making it very difficult for a taxonomist task.
Another fundamental reason is that a large fraction of protists are unculturable, meaning they cannot be grown in a laboratory, hampering their study by scientists.
One such enigmatic uncultured lineage is named ‘MASTs’ (MArine STramenopiles). They represent a major community among aquatic protists. Considered as heterotrophs, they secrete enzymes that digest food externally after grazing on bacteria (phagocytosis). In fact, these tiny protists (up to 3µm) consume efficiently viruses, bacteria, picophytoplankton and are ingested mainly by other protists and small crustaceans. They are particularly adapted to this process as they possess special appendages, a long whip-like structure, used to create feeding currents that enhance prey capture. These heterotrophic protists are still hugely understudied and so cause a real taxonomic gap in our knowledge of the earth’s biodiversity.
In response, new technologies have facilitated genome investigation with the development of new techniques in environmental molecular sequencing. Single cell genomic analyses represent a broad and fast solution to build more reference databases for novel and uncultured species. In single-cell genomics, an individual microbial cell is isolated, lysed and the whole genome is amplified. By studying entire genomes rather than individual genes, single-cell contributions have a great potential to understand the full complexity of cells for novel microbes (suffering from a real lack of reference protist genomes) by revealing genome-wide properties to explore cell identity.
In this context, we aim to investigate ecological relationships and trophic role within the plankton community. However, the lack of systematic genome comparisons of marine microorganisms leads to a lack of understanding of marine biology in general. Hence, we would provide an effort in sequencing more heterotrophic flagellates with a focus on the MArine STramenopiles, which we believe will lead to a model for investigating links between diversity and ecological function in marine systems. Since the MASTs are divided into 18 sub-groups with the potential for associated ecological niches for each, recovering a large part of the MASTs distribution over their “family tree” will fill this gap in under-sampled areas and increase the taxonomic heterogeneity.
In pursuit of reconstructing total eukaryotic biodiversity, better efforts must be made to sample the whole of the ocean’s biodiversity by developing new techniques and methods. Among these are i) the power of collaborative open research and ii) increasing the general public’s interest, allowing them to perceive the crucial role of protists in our oceanic ecosystems.
About the author

Aurelie Labarre – ESR 1
I started my academic training in bioinformatics and genomics as a Bachelor and a graduate student at the University of Laval in Canada. A key part of my graduate work was studying the genomic evolution of eukaryotic micro-organisms within the Chlorophyceae lineage. To gain more direct work and research experience, I then moved to the UK to take part in a bioinformatics genome project of a Oomycete-related organism; followed by a metagenomic project in Norway in order to broaden my experimental and analytical skills. Through all these experiences, I have been fascinated by the world’s rich biodiversity and my progress in these research areas has inspired me to want to work more on the genomic diversity of marine protists.


Leave a Reply
Want to join the discussion?Feel free to contribute!