SINGEK: New paper about SCG of oceanic unculturable picoeukaryotes
We are glad to announce that a new SINGEK paper is out in Scientific Reports/ NatureJournal:
“Accessing the genomic information of unculturable oceanic picoeukaryotes by combining multiple single cells”
J-F. Mangot, R. Logares, P. Sánchez, F. Latorre, Y. Seeleuthner, S. Mondy, M. E. Sieracki, O. Jaillon, P. Wincker, C. de Vargas and R. Massana
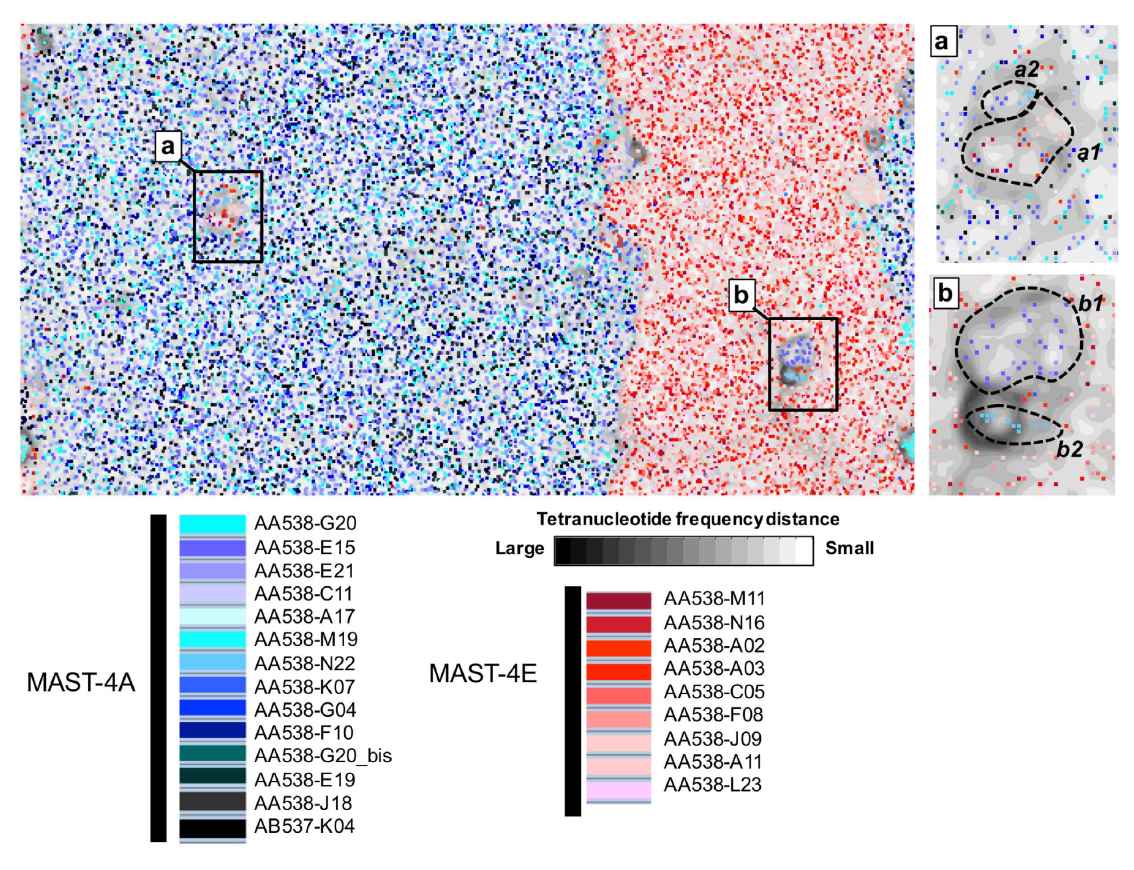
Comparison of tetranucleotide frequencies of SAGs in an ESOM map.
Pico-sized eukaryotes play key roles in the functioning of marine ecosystems, but we still have a limited knowledge on their ecology and evolution. The MAST-4 lineage is of particular interest, since it is widespread in surface oceans, presents ecotypic differentiation and has defied culturing efforts so far. Single cell genomics (SCG) are promising tools to retrieve genomic information from these uncultured organisms. However, SCG are based on whole genome amplification, which normally introduces amplification biases that limit the amount of genomic data retrieved from a single cell. Here, we increase the recovery of genomic information from two MAST-4 lineages by co-assembling short reads from multiple Single Amplified Genomes (SAGs) belonging to evolutionary closely related cells. We found that complementary genomic information is retrieved from different SAGs, generating coassembly that features >74% of genome recovery, against about 20% when assembled individually. Even though this approach is not aimed at generating high-quality draft genomes, it allows accessing to the genomic information of microbes that would otherwise remain unreachable. Since most of the picoeukaryotes still remain uncultured, our work serves as a proof-of-concept that can be applied to other taxa in order to extract genomic data and address new ecological and evolutionary questions.